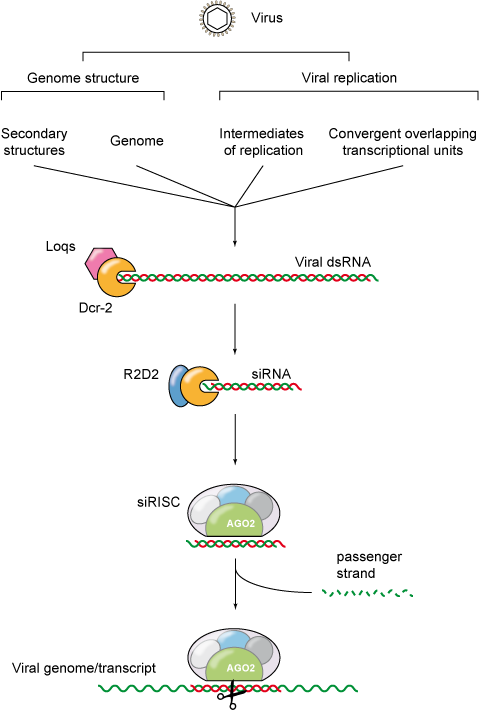
Figure 1: RNA silencing against virus infection.

Research
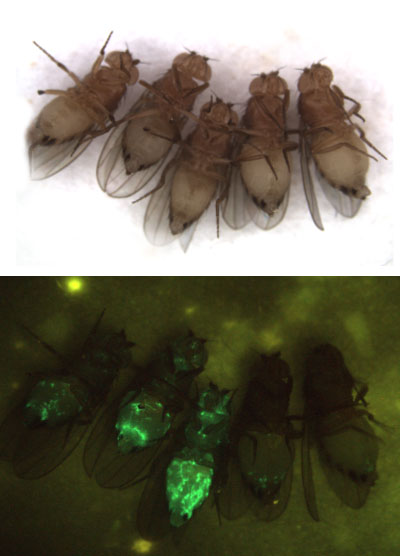
Figure 2: Our model system, the fruit fly Drosophila melanogaster. Top panel: ventral view of 5 female flies. Bottom panel: the same flies infected with Sindbis virus (arbovirus) expressing a green fluorescent protein. The first fly on the right is non infected. The virus infection is observed as a green stain throughout the body with a higher replication level in the fat body.
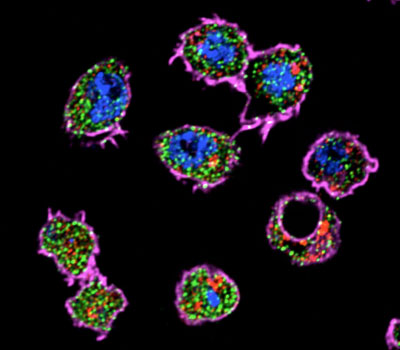
Figure 3: AGO2 is enriched in the endosome fraction of Drosophila S2 cells (plasma membrane_actin = magenta, nucleus = blue, Ago2 = green, late endosomes_Rab7= red).


If you are not a scientist, here’s a simple summary of our research:
Insects have an immune system that allows them to live undisturbed when they are infected by a virus that is deadly to human beings. We are trying to understand how this immunity operates.
It is complicated, expensive and very difficult to work with mosquitoes in order to understand the transmission of viruses from insects to humans. Lucky for us, fruit flies have the same immune system as mosquitoes. They are easier to handle and cultivate (no blood meals required) and universally accepted as a model system for studying insect immunity. So we study fruit flies under high-powered microscopes, allowing us to infect them and then dissect their immune system.
If you are a scientist, here’s a more scientific explanation of our research:
The overall goal of our research is to understand the molecular and cellular aspects underlying the immune dynamics of the antiviral response in insects. We intend to turn the focus from the now well-studied field of antimicrobial immunity towards the less-studied field of antiviral immunity. Very few laboratories in the world have united the experimental and bioinformatic approaches that we have succeeded in combining over the past years. We believe, therefore, that our scientific strategy offers new perspectives on emerging viral disease transmission and will inspire a new way of thinking about immunity.
RNA interference and viral immunity in insects
RNA interference (RNAi) is a conserved sequence-specific gene-silencing mechanism that is induced by double-stranded RNA (dsRNA). One of the functions of this pathway is the defense against parasitic nucleic acids such as transposons or viruses.
Double-stranded RNA is commonly generated during viral replication and is considered to be a molecular pattern characteristic of viral infection. Viral dsRNA originates from the viral genome itself (dsRNA viruses and secondary structures), from viral intermediates of replication (RNA viruses with exception of the retroviruses), or from convergent overlapping transcriptional units (DNA viruses). Thus, dsRNA is often a signature of virus infection, and as such it triggers a range of host responses, such as RNA interference (RNAi).
When RNAi was shown to be antiviral, first in plants in 1997 and later in nematodes and in insects, a new and intriguing phase opened up in host-pathogen relationships and immunity: a new “nucleic-acid based” immune system was discovered and still awaits to be unravelled.
Important viral infectious diseases, such as Dengue and Chikungunya, are transmitted to humans by insect vectors. One of the key factors that modulates whether an insect is competent or not to transmit a given pathogen is its innate immune response. Arboviruses (for arthropod-borne viruses) are maintained in a transmission cycle between hematophagous insect vectors (and arthropods in general), and vertebrate hosts. Within their insect vector, arboviruses encounter several anatomical and immunological barriers, of which RNA interference (RNAi) is the major antiviral defence mechanism. The small interfering RNA (siRNA) pathway constitutes a hallmark of the insect antiviral RNAi response. During the siRNA response, the defence mechanism is activated by cleavage of viral double-stranded RNA (dsRNA) into 21 nucleotide (nt) viral small interfering RNAs (vsiRNA) by Dicer-2 (Dcr-2). Once produced, vsiRNAs guide the sequence-specific recognition and cleavage of viral target RNAs by an Argonaute-2 (AGO-2)-containing RNA induced silencing complex (RISC). It was recently proposed that another RNAi pathway is involved in antiviral defence: the piwi-interacting RNA (piRNA) pathway. Indeed, we showed that mosquito cells infected with (+) and (-) RNA arboviruses produce viral piRNAs (vpiRNA) with the hallmarks of ping-pong amplification, a broader size range (25-27 nts) and associated to PIWI proteins of the Argonaute family. These observations suggest that the piRNA pathway constitutes another facet of the antiviral RNAi response in insects that could act in combination with the already-described siRNA pathway to control viral infection.
An ideal model system to study antiviral immunity and host-pathogen relationships should combine a genetically tractable small animal with a virus capable of naturally infecting it. Using Drosophila and an array of viruses infecting it as a model system, we have performed work key to redefine and understand better the principles of the antiviral RNAi response:
- We obtained evidence that the antiviral defense mechanism in Drosophila melanogaster is critically dependent on components of the RNAi machinery.
- We found that dsRNA reaches the RNAi machinery via a specific pathway that involves clathrin-mediated endocytosis and scavenger-like pattern-recognition receptors.
- We showed that antiviral immunity in Drosophila requires systemic RNA interference spread.
- We established that RNAi is the main antiviral response against (+) and (-) strand RNA viruses and DNA viruses in insects.
- We contributed to the exploitation of high throughput sequencing data by developing software for in silico reconstruction of viral genomes from small RNAs.
- We showed the production of viral piRNAs in infected mosquito cells.
- We discovered how persistent viral infections are established in the insect model Drosophila by the interplay of RNAi and retrotransposition.
To consolidate the on-going projects in the laboratory and the understanding of antiviral RNAi, we are beginning to explore new areas of RNAi antiviral immunity using an interdisciplinary approach: Drosophila genetics and mosquitoes to carry out most of the experiments, as well as concepts and techniques from classical and molecular virology, immunology, biochemistry and cell biology. As you would expect, next-generation sequencing and state-of-the-art bioinformatic tools are integral parts of our experiment strategy.
Currently we are pursuing the following projects:
- Dissecting dsRNA uptake in RNAi-based antiviral immunity in vivo.
- Exploring the role of different Drosophila proteins in the spread of the RNAi immune signal throughout the body.
- Studying the role of viral piRNAs on RNAi-mediated antiviral response.
- Deciphering the mechanism of viral persistence and transmission in insects.
- Ascertaining the existence of long lasting immunity in insects.
- Measuring the genetic interaction between virus and insect innate immunity during the course of natural infection.